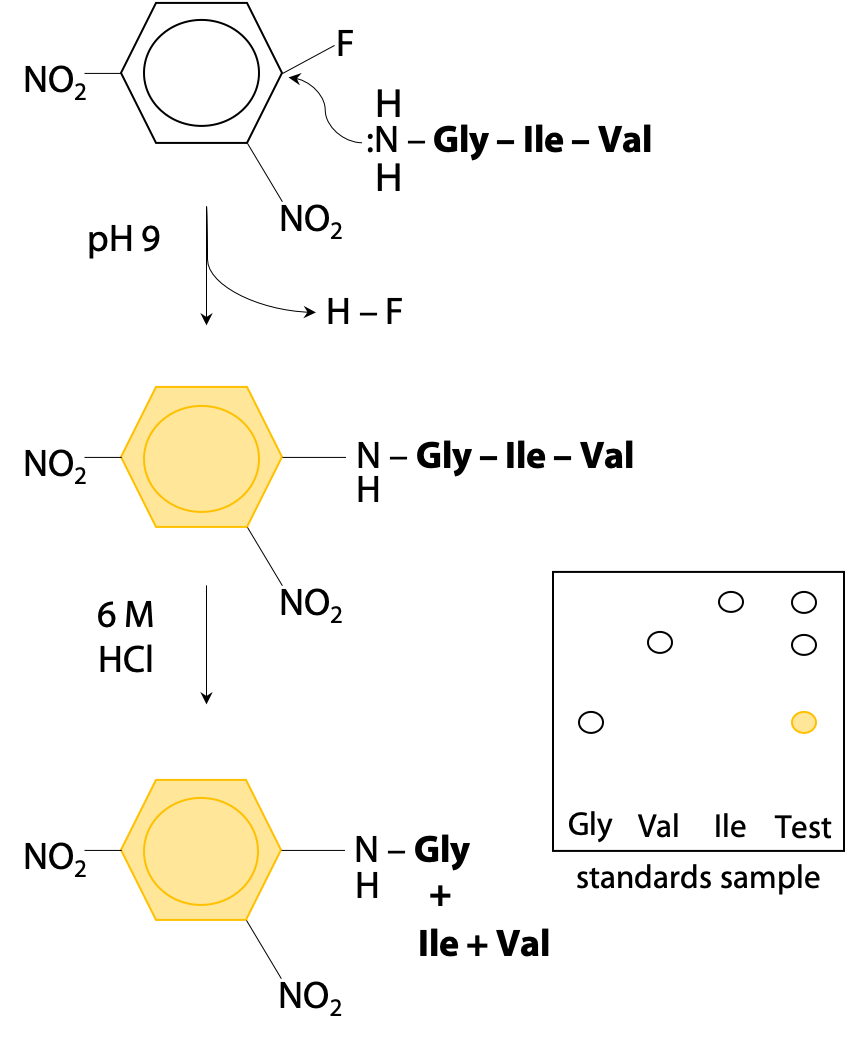
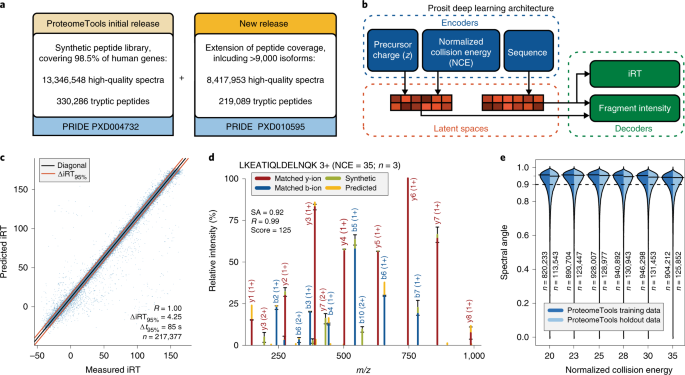
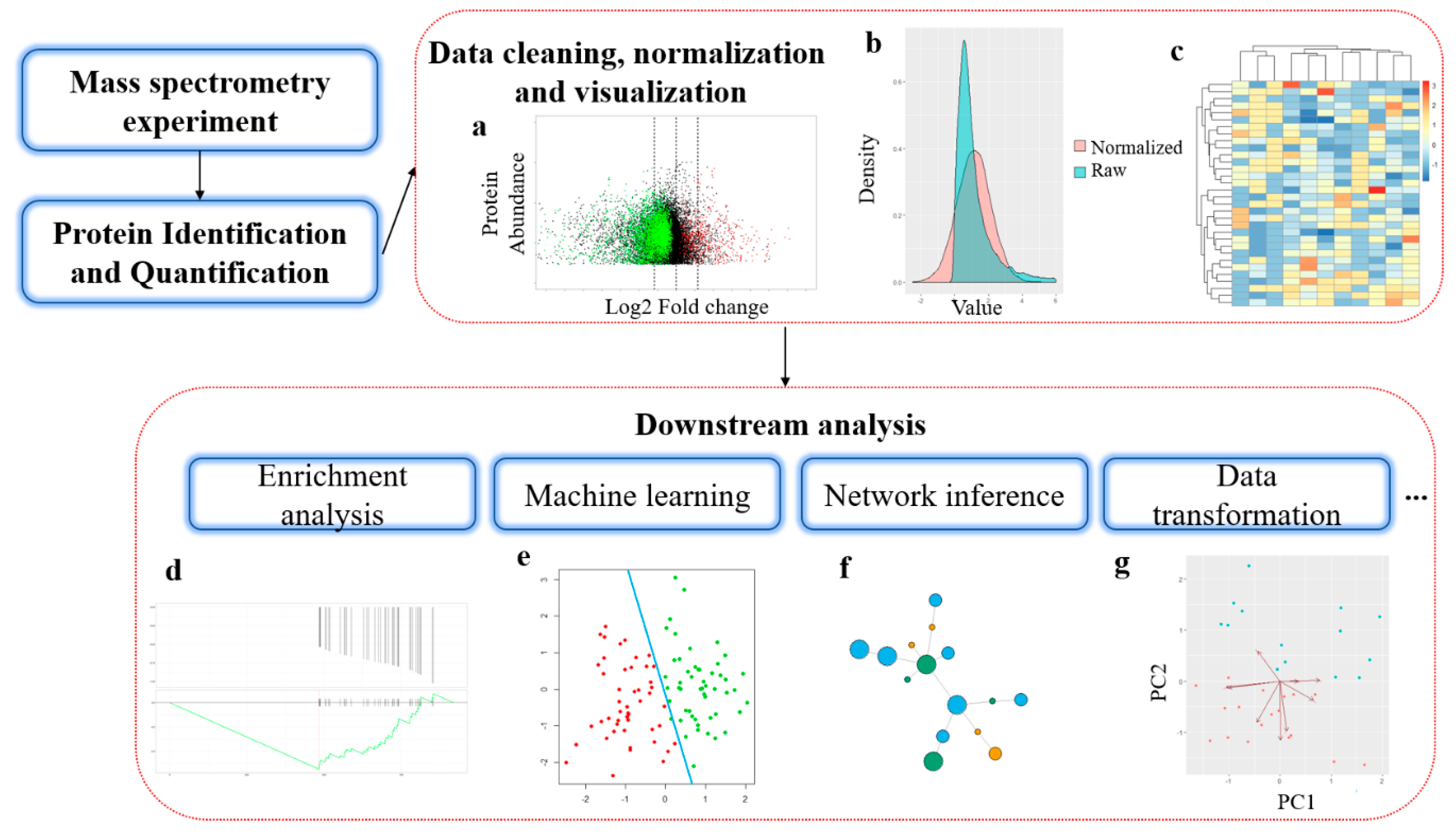
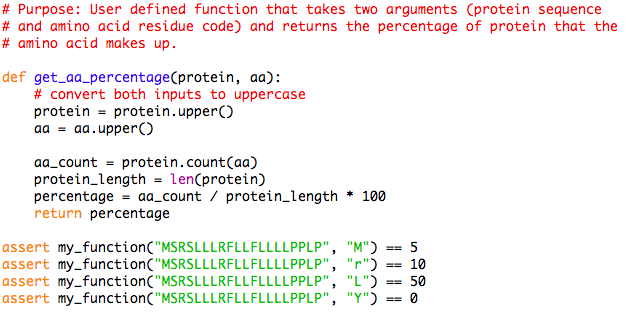
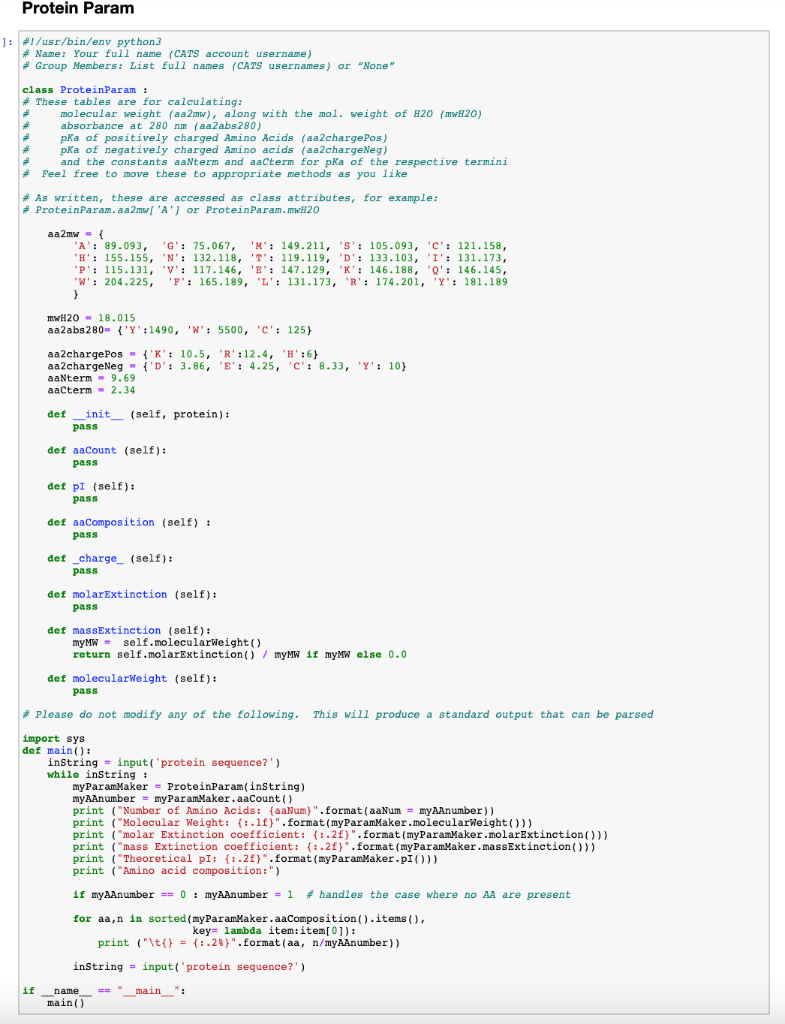
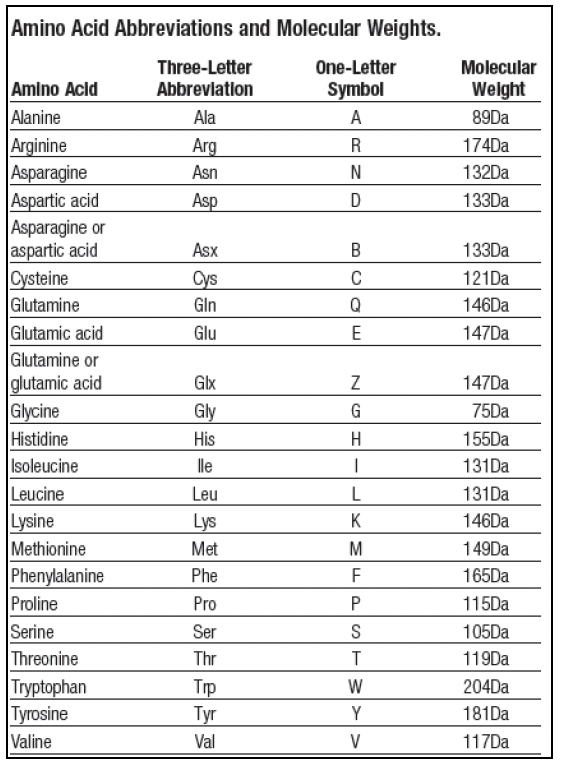
GitHub - efr-essakhan/pepfeature: A Python package that has functions for featue calculation of protein sequences. Features that are calculated by this package are niched for Epitope prediction.
BioPython_Scripts/HowTo_Translate_DNA_fasta_to_Protein_Fasta.txt at master · PNNL-Comp-Mass-Spec/BioPython_Scripts · GitHub
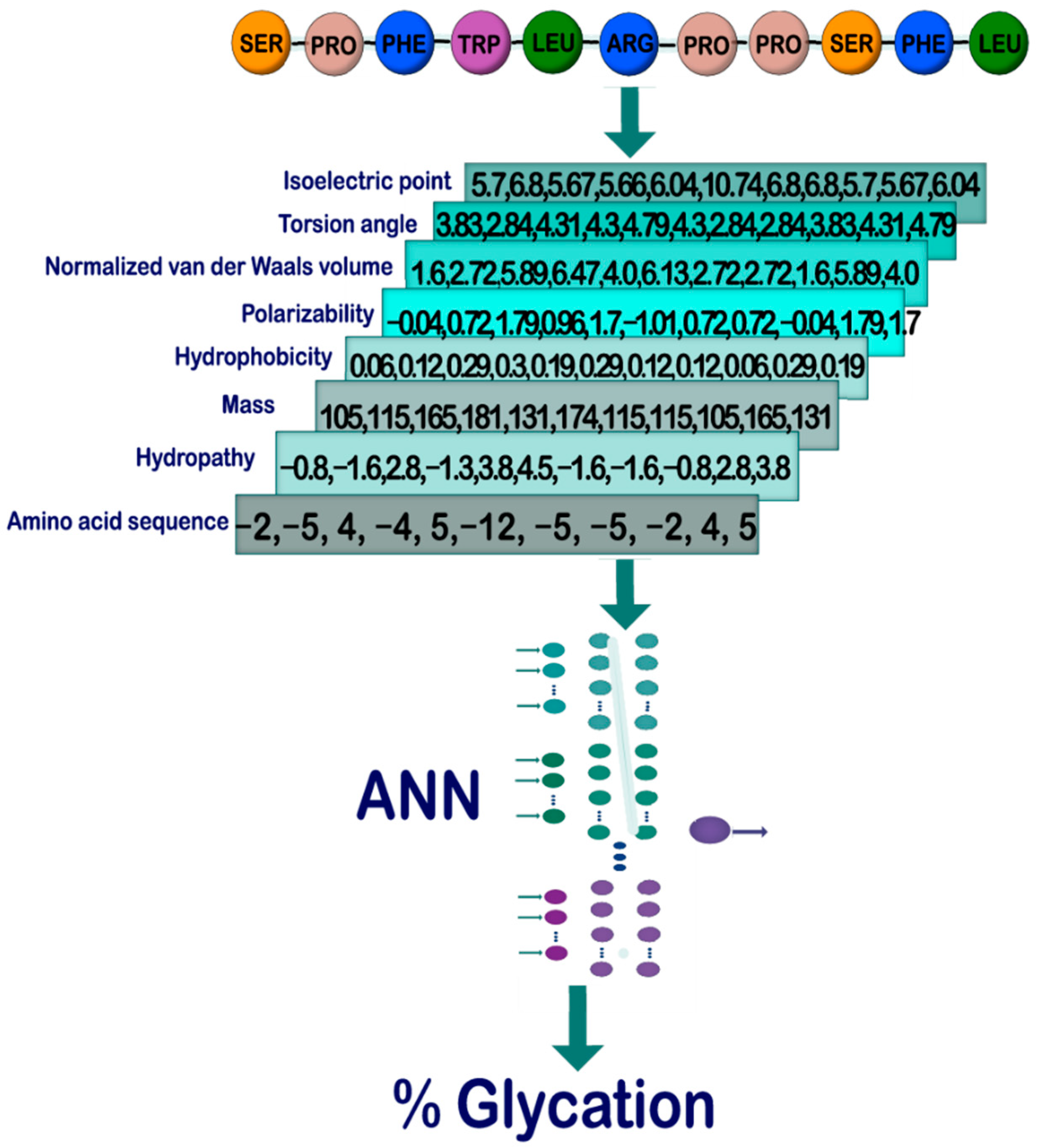
Sensors | Free Full-Text | On the Prediction of In Vitro Arginine Glycation of Short Peptides Using Artificial Neural Networks
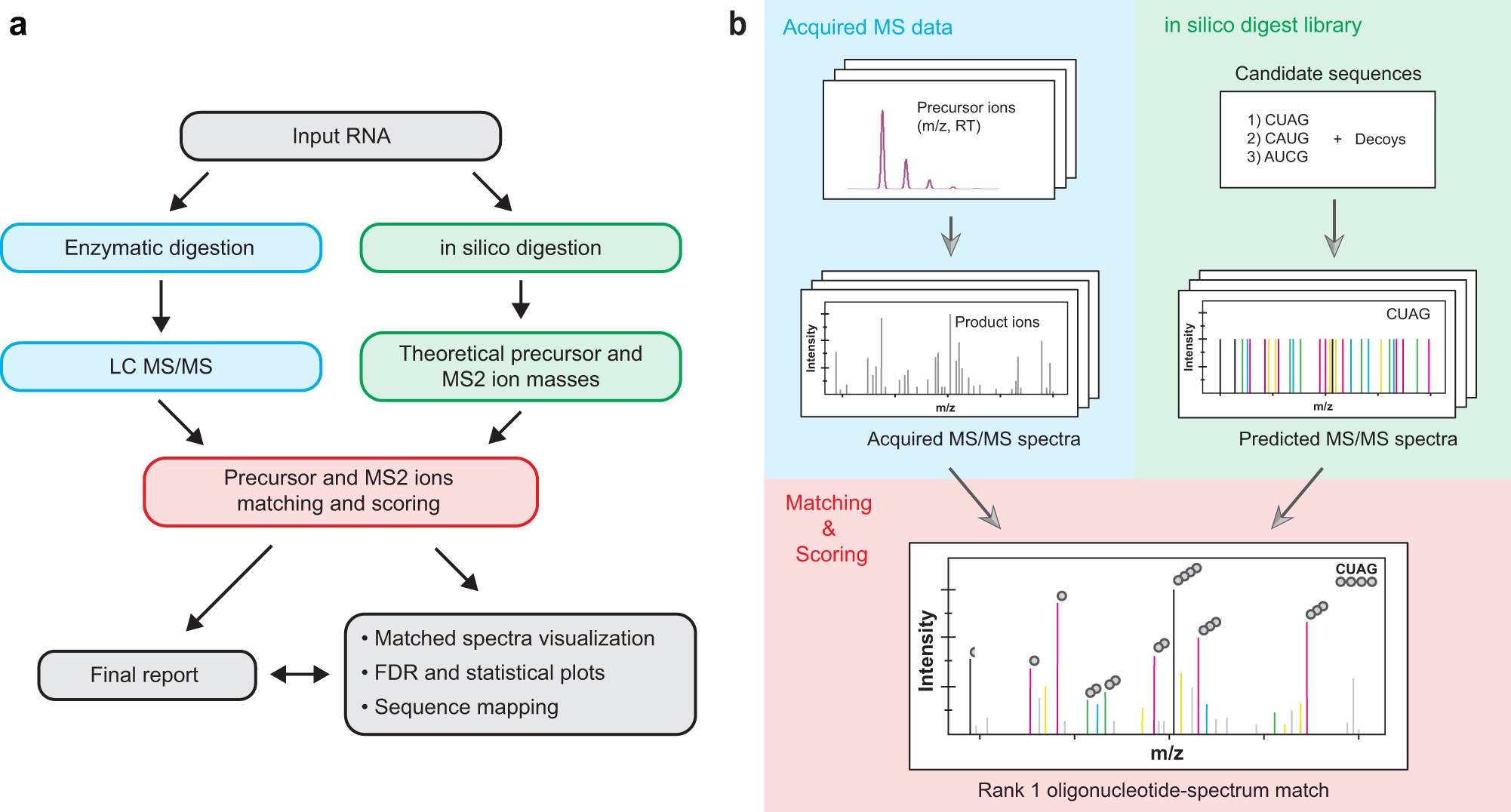
Pytheas: a software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry | Nature Communications
ProteoClade: A taxonomic toolkit for multi-species and metaproteomic analysis | PLOS Computational Biology
Functional expression of diverse post-translational peptide-modifying enzymes in Escherichia coli under uniform expression and purification conditions | PLOS ONE